This manual covers use of ZEN 2.3 Pro on the Live Cell Station.
Before turning on the system, first make sure the slide adapter is already installed on the stage. If not, install it before powering on the system. If the microscope is already on, switch off the microscope (#4) before the slide adapter is installed.
- If you will be using fluorescence turn on the Xcite lamp (#1), otherwise leave it off
- Power strip on top shelf (#2)
- Microscope (silver push switch on back left, #4)
- (Optional) Evolve EMCCD camera (#5), if you are going to use it
- Start up computer (#6), log in the computer and start ZEN2.3
The Microscope
There are 4 stage inserts for holding different samples - a heated insert for 35 mm glass-bottomed dishes, an insert for multi-well plates, a universal frame for holding slides and dishes, and a universal frame with clamps for holding slides. The last one is preferred for tile imaging.
Nearly everything on the microscope is motorized so can be controlled either on the LCD touchscreen or through ZEN.
Objectives
|
Mag
|
NA
|
Oil?
|
DIC?
|
|
5x
|
0.16
|
NO
|
no
|
|
10x
|
0.30
|
NO
|
Ph1
|
|
20x
|
0.8
|
NO
|
DIC
|
|
40x
|
0.75
|
NO
|
DIC
|
|
63x
|
1.4
|
Yes
|
DIC
|
|
100x
|
1.4
|
Yes
|
DIC
|
When ZEN starts, the 'Locate' tab is opened by default. Lower down the objective a bit before loading a slide; Position the sample right above the objective
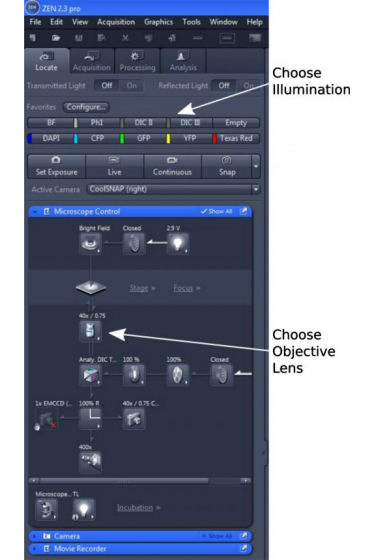
- Choose the objective lens you wish to use (on touch pad, in 'Locate', or from the objective box on the right side of ZEN window)
- Select an illumination you want to use. The quick access buttons are available for BF, PH1, DICII, DICIII, DAPI, CFP, GFP. YFP, and Texas Red. Cy5 (Far Red) and Cy7 (near-infrared) filter cubes are not installed, but are available when requested.
- Coarse/fine focus knobs are on the microscope. X-Y stage movement is through the rotary controller;
- Find and focus on the sample
- Click "Acquisition" tab (step 1)
- Choose a presetting that matches the camera you wish to use (Evolve EMCCD attached to the left or CoolSNAP ES2 to the right of the microscope) (step 2)
- Tick all the channels you wish to image (step 3)
- Highlight a channel and enter a relatively short exposure time (step 4); Don't use 'Auto Exposure' or 'Set Exposure'
- Start 'Live' (step 5); A live image shall show up in the 'center screen area'.
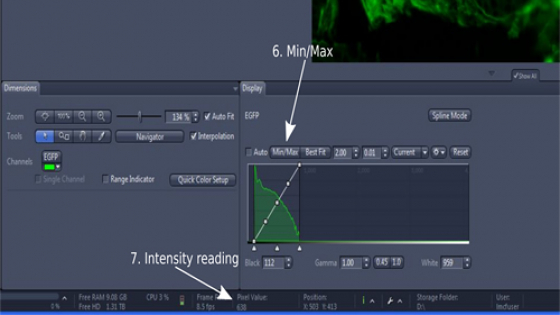
- Click 'Min/Max' or tick 'auto' in 'Display' section right below the image (step 6); This will adjust the dynamic range of the image for easy viewing of it.
- Hover the mouse pointer over your interested objects and read the intensity ('Pixel Value' in the 'Status bar' at the bottom of the program window (step 7); Read also the intensity of a clean background; Adjust exposure time and/or excitation light intensity so that the intensity difference is 3 times or more for a publication quality image; But you need to watch out for oversaturation (histogram touches the right boundary) and photobleaching of the sample due to excess exposure.
- Repeat steps 4-7 for all other channels
- Click 'Snap' to take images for all selected channels (step 8)
- After acquisition, a merged image is displayed. Individual channels can be separately displayed by click 'Split'.
- All acquired images will be lined up on the right side of the ZEN window; Save images in '.czi' format; Close saved images to free up the computer memory
Tiling and stitching is used to generate a high-resolution image that covers an larger area of multiple continuous fields.
A. Quick tile image acquisition
You need to find a proper imaging condition for each channel (see above) before tile scanning and stitching. This function is built in 'Snap' button. You can choose to 'Snap' single image (see above), 2x2, 3x3, 4x4, or 5x5 image tiles around the current position.
B. Standard tile image acquisition
- Set up exposure for all channels as described above
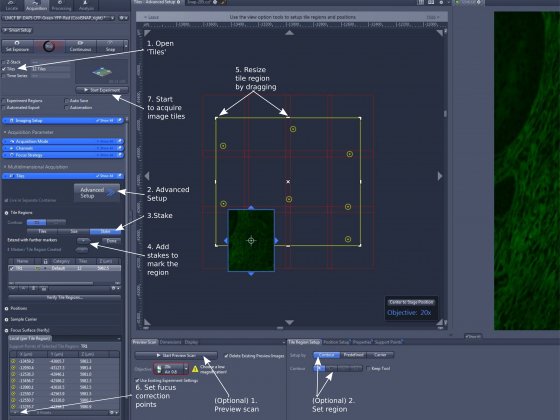
- Open the functional module 'Tiles' (step 1)
- Open 'Advanced Setup' (step 2) and the 'Live' function starts. The center screen area is split into two regions: a vitual stage on the left and a live image of the current field on the right.
- Mark the area to be imaged ('Tile Regions'):
- Highlight 'Tiles' or 'Stakes (step 3).
- To mark region by 'Tiles' simply put numbers in row and column, and click plus sign (+)
- To mark region by 'Stakes', move the stage manually using the rotary controller or by double-clicking in the virtual stage panel in ZEN, add stakes (step 4); You need to put down at least two 'stakes' for the software to mark an area around them (marked with yellow borders). The actual area that the final image covers shows up as red colored tiles.
- Moving the stage to a position outside the current region (red lined) and adding 'stake' again will expand the boundary. You can also drag the yellow line with the mouse to resize the area (expanding or trimming) (step 5 ).
- (Optional) Tile region setup following a preview scan:
- Set up a region (see above) using a low magnification (e.g. 5x) objective
- "Start Preview Scan" (optional step 1)
- In 'Tile Region Setup', choose a desired 'Contour' (e.g. rectangle, oval, free-drawing) (optional step 2) and draw a region in the previewed image; The newly marked area will show up as a new tile region in the region list.
- Change to objective you wish to use and set focus correction points (see below)
- Set focus correction points ('Focus Surface'): this function allows the program to use focuses determined by the user to counteract the uneven surface of the tissues section.
Move the stage to a position, adjust focus, and add ('+') the position (X, Y, and Z) in memory (step 6); Repeat adding focus correction points; You don't have to set focus for all tiles. For image tiles without focus correction points, the software will calculate the focuses based on other focus points.
- 'Start Experiment' to take the image tiles (step 7).
- Stitching: to have the image tiles stitched as a single image
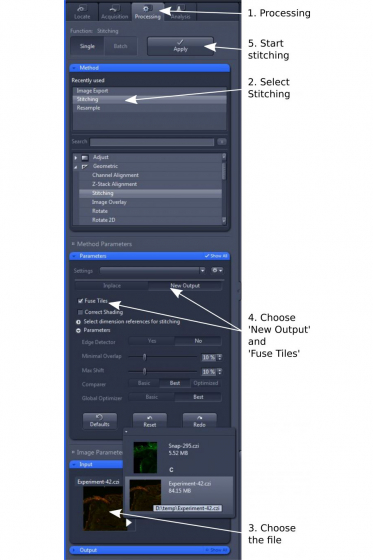
- Open 'Processing' tab and select 'Stitching' (step 1 and 2)
- In 'Input', choose the image tiles you wish to stitch (step 3)
- In 'Parameters', choose 'New Output' and tick 'Fuse Tiles' (step 4); Other parameters are mostly default, but can be adjusted for best alignment and stitching.
- Click 'Apply' to start the stitching process (step 5). The stitched image will be displayed as a separate image.
- (Optional) Making image from region of interest (ROI): You may need to display or publish a portion of the stitched image, or you just want to cut out the blank areas near the edges to reduce the file size; Perform this function only on stitched images.
- Draw a 'region of interest' in the image using the 'ROI' tool under 'Graphics'
- Right-click in the marked area; Click 'Create Subset Images from Region of Interest' to duplicate the image within selected region
- (Optional) 'Resample' the image: The stitched images are usually big in file size. There are chances that you want to reduce the size of the files by 'resampling' - scaling down:
- Open 'Processing' tab and select 'Resample'
- In 'Input', choose the image
- In 'Parameters', put in a 'Scaling' factor that is smaller than 1 for both X and Y axises
- 'Apply'
- Save images in '.czi' format. Close saved images to free up the computer memory
A. Contrast adjustment before image export:
The acquired image is shown as channel-merged. 'Split' the channels to show individual channels.
In 'Display', click the channel to be ajusted (step #1), drag the triangle under the histogram for white and black points to proper positions (step #2), so that the main structure is lightened up but not oversaturated and the background is dark. Do the contrast adjustment in a sensitive and reasonable way. Usually, the triangle for black point needs to be positioned right inside the main curve of the histogram. You can also double-click on a channel to have only the channel displayed, then hover the mouse cursor in the background area, read the intensity of the pixels (under 'Pixel Value'), type in the number for black cut-off point (step #3).
Please keep in mind, adjustment of the contrast only affects the display of the image. It does not change the intrinsic intensity values of the image.
B. Image export:
Under 'Method' in 'Processing', choose 'Image Export', in 'Input' choose the image to be exported, in 'Parameters', use TIFF as file type (if a stitched file is too big, JPEG is an acceptable type too), tick 'Apply display Curve and Channel Color', 'Burn-in Graphics', 'Merged Channels Image', and 'Individual Channels Image' if you wish the exported image will look as displayed, then 'Apply' to let the image exported
Check (or re-check) the booking calendar online; If somebody will use the scope after you in one hour, clean the oil objective lens with lens tissue if you have used it, close ZEN, leave the system on; If the person after you is not going to use the fluorescent light source (#1), turn it off; If the next user does not indicate whether he or she will use the fluorescent light, leave it on
When nobody comes to use the system in one hour, turn the system off in the backward order of startup::
- Close ZEN, shut down the computer (#6)
- Evolve EMCCD camera (#5) if you have used it
- Microscope (#4)
- Power strip (#2)
- Fluorescent light source (#1)